Owner Yann: (Experimental) Demo Google cloud application flow for covid19 data extraction from PDF's
|
|
5 vuotta sitten | |
|---|---|---|
| content | 5 vuotta sitten | |
| notebooks | 5 vuotta sitten | |
| scripts | 5 vuotta sitten | |
| .DS_Store | 5 vuotta sitten | |
| README.md | 5 vuotta sitten | |
| requirements.txt | 5 vuotta sitten |
README.md
COVID-19 public dataset on GCP from cases in Italy
Medical notes and entities from TRUE patient cases publicly available on BigQuery and Datastore!
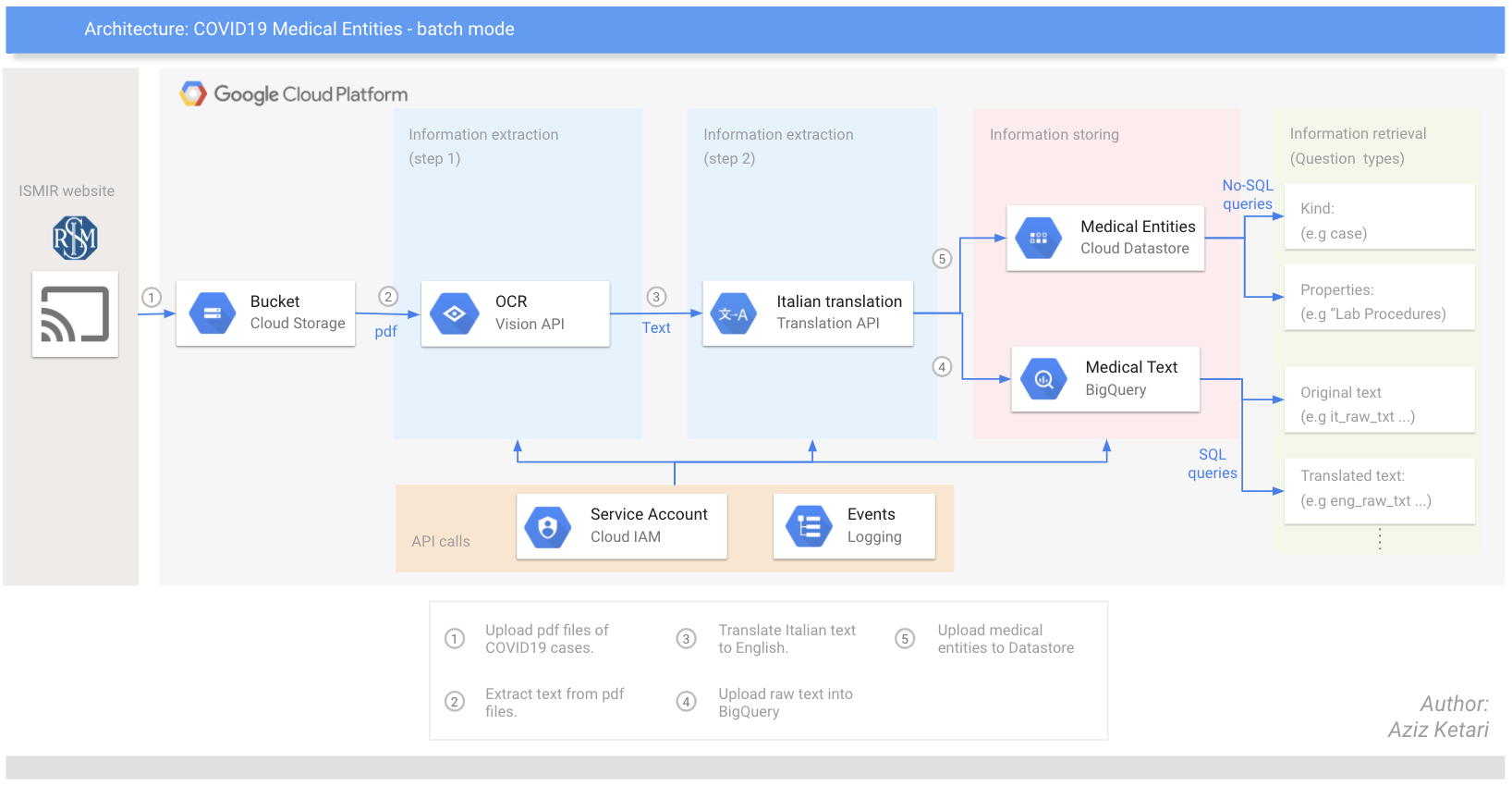
This repository contains all the code required to extract relevant information from pdf documents published by ISMIR and store raw data in a relational database and entities in a No-SQL database.
In particular, you will use Google Cloud Vision API and Translation API, before storing the information on BigQuery. Separately, you will also use specific NER models (from Scispacy) to extract (medical) domain specific entities and store them in a NoSQL db (namely Datastore) on Google Cloud Platform.
Looking for more context behind this dataset? Check out this article.
Google Cloud Architecture of the pipeline:
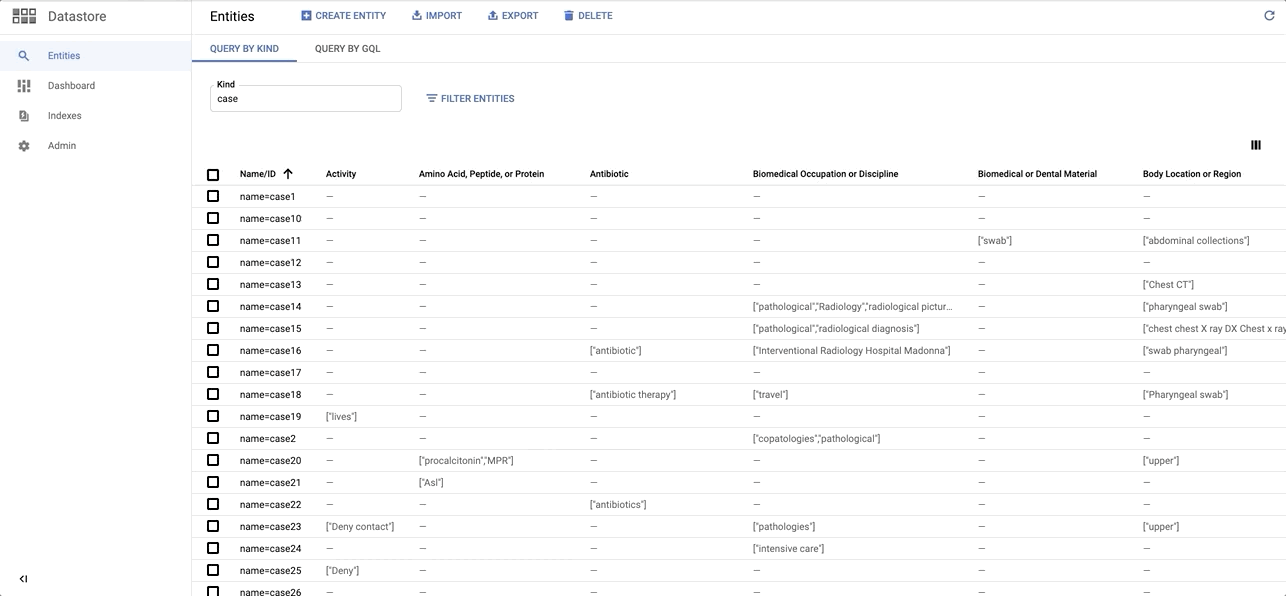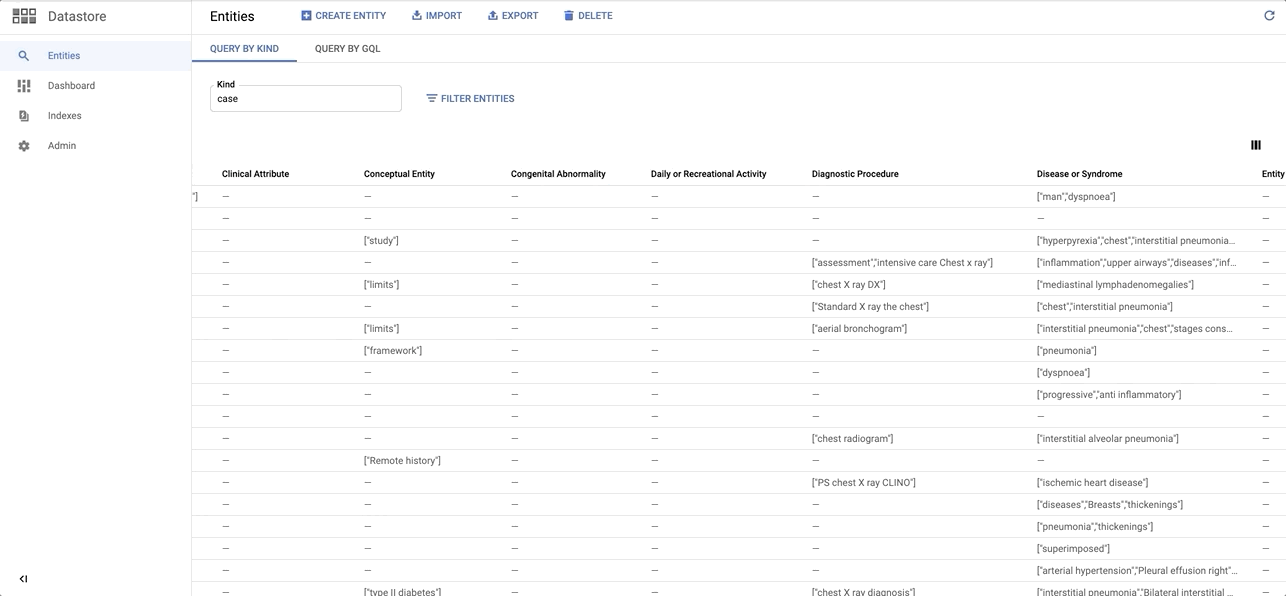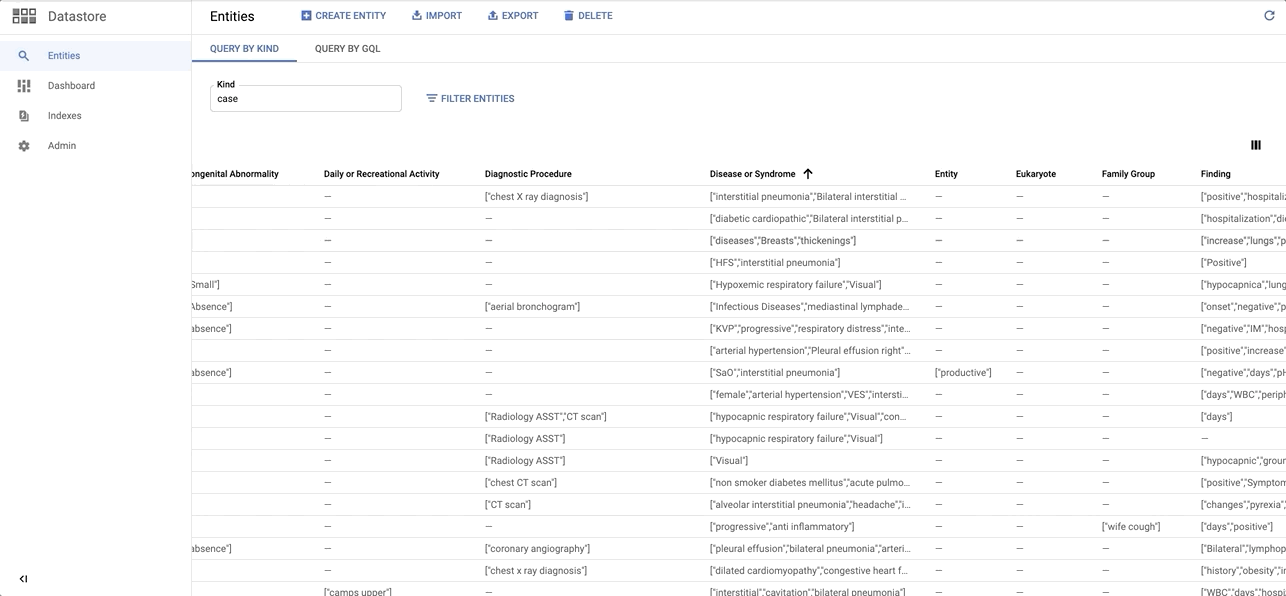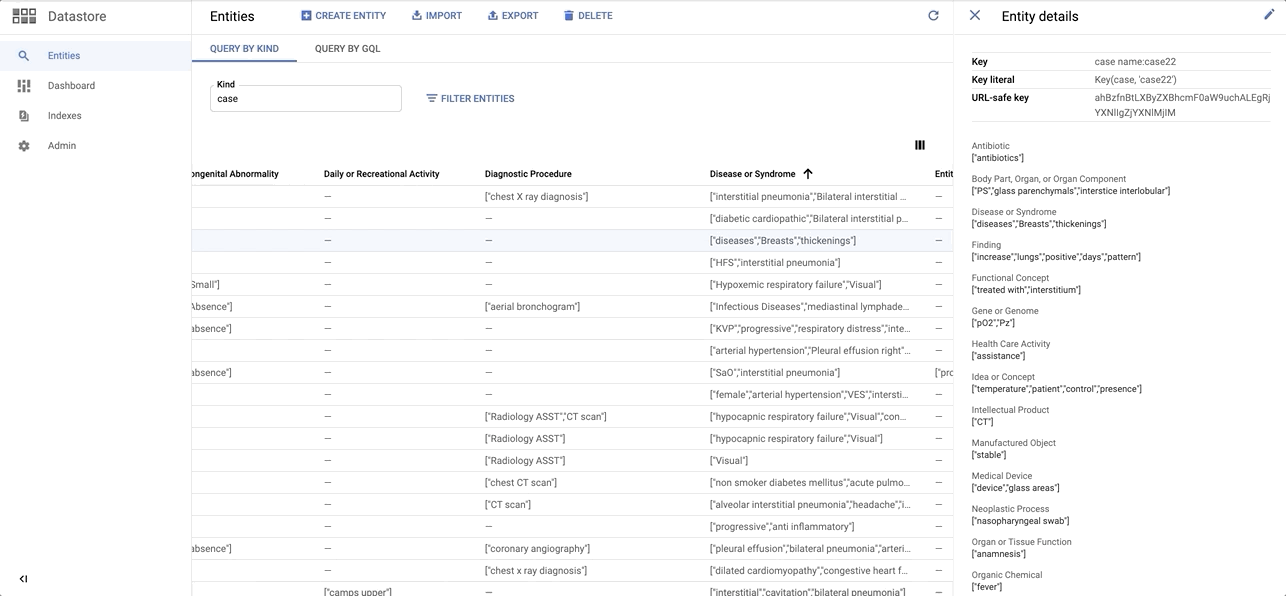
Quick sneak peak on the Entity dataset on Datastore:
Installation
You can replicate this pipeline directly on your local machine or on the cloud shell on GCP.
Requirements:
- Clone this repo to your local machine using https://github.com/azizketari/covid19_ISMIR.git
- You need a Google Cloud project and IAM rights to create service accounts.
- Create and Download the json key associated with your Service Account. Useful link
Modify the values to each variables in env_variables.sh file then run
cd ./covid19_ISMIR source ./content/env_variables.shSet the project that you will be working on:
gcloud config set project $PROJECT_ID
Enable APIs:
gcloud services enable vision.googleapis.com gcloud services enable translate.googleapis.com gcloud services enable datastore.googleapis.com gcloud services enable bigquery.googleapis.comInstall package requirements:
Make sure you have a python version >=3.6.0. Otherwise you will face some version errors Useful link
ERROR: Package 'scispacy' requires a different Python: 3.5.3 not in '>=3.6.0'
pip3 install --user -r requirements.txt
Note:
You will also need to download a Named Entity Recognition model for the second part of this pipeline. See Scispacy full selection of available models here. If you follow this installation guide, the steps will automatically download a model for you and install it.
Extracting data
Step 1: Download the required files to your bucket and load the required model in your local
(this step will take ~10 min)Optional: If you have already downloaded the scispacy models, you should modify the file ./content/download_content.sh to not repeat that step
source ./content/download_content.shStep 2: Start the extraction of text from the pdf documents
python3 ./scripts/extraction.py
Pre-processing data
Following the extraction of text, it's time to translate it from Italian to English and curate it.
python3 ./scripts/preprocessing.py
Storing data
Following the pre-processing, it's time to store the data in a more searchable format: a data warehouse - BigQuery - for the text, and a No-SQL database - Datastore - for the (UMLS) medical entities.
python3 ./scripts/storing.py True True [Model_of_your_choice]
Test
Last but not least, this script will run a few test cases and display the results. Feel free to modify the test cases.
python3 ./scripts/retrieving.py
Contributing
To get started...
Step 1
Option 1
- 🍴 Fork this repo!
- 🍴 Fork this repo!
Option 2
- 👯 Clone this repo to your local machine using https://github.com/azizketari/covid19_ISMIR.git
- 👯 Clone this repo to your local machine using https://github.com/azizketari/covid19_ISMIR.git
Step 2
- HACK AWAY! 🔨🔨🔨
Step 3
- 🔃 Create a new pull request
