14 измененных файлов с 15 добавлено и 13 удалено
BIN
.DS_Store
+ 9
- 7
README.md
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
BIN
data/.DS_Store → content/.DS_Store
+ 0
- 0
data/UMLS_tuis.csv → content/UMLS_tuis.csv
+ 0
- 0
data/download_content.sh → content/download_content.sh
+ 0
- 0
data/images/.DS_Store → content/images/.DS_Store
+ 0
- 0
data/images/bq_snapshot.gif → content/images/bq_snapshot.gif
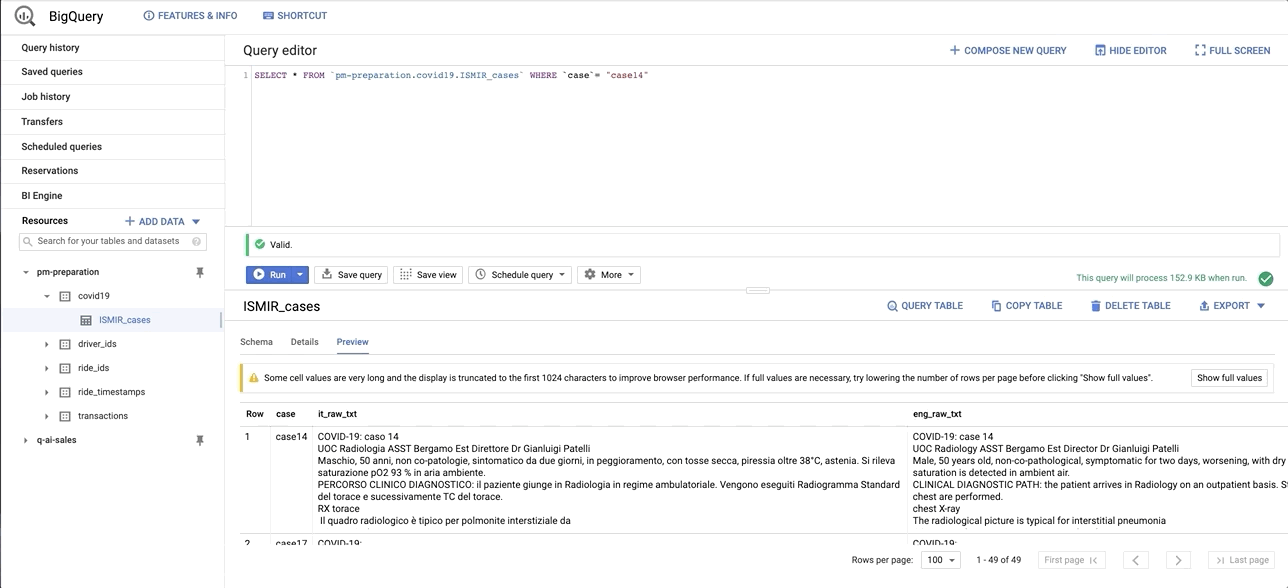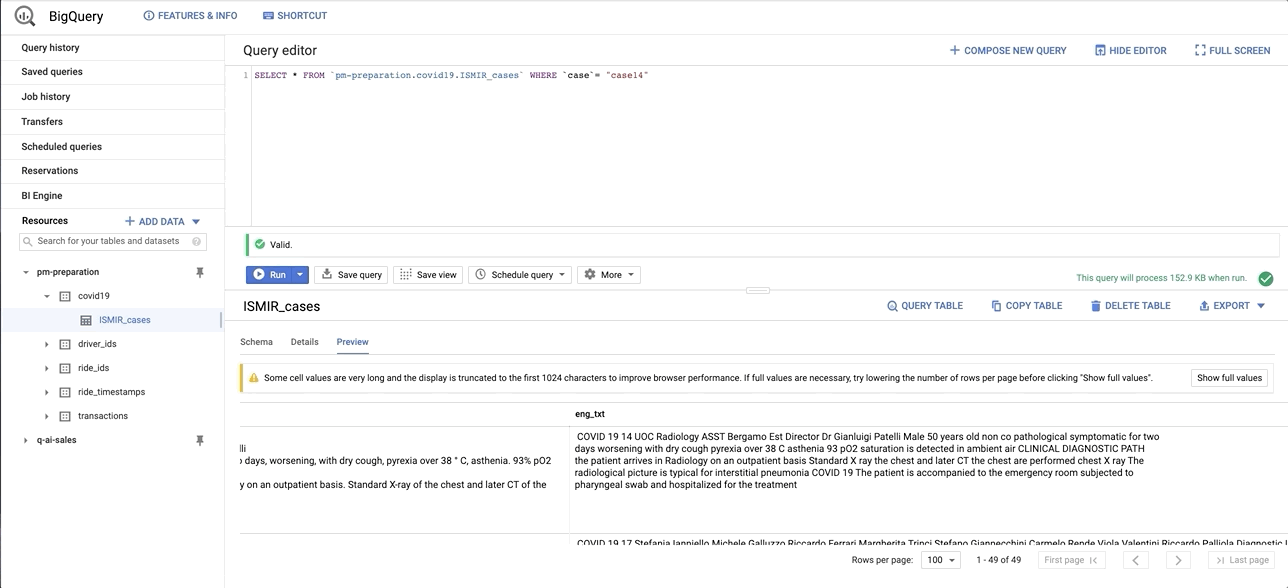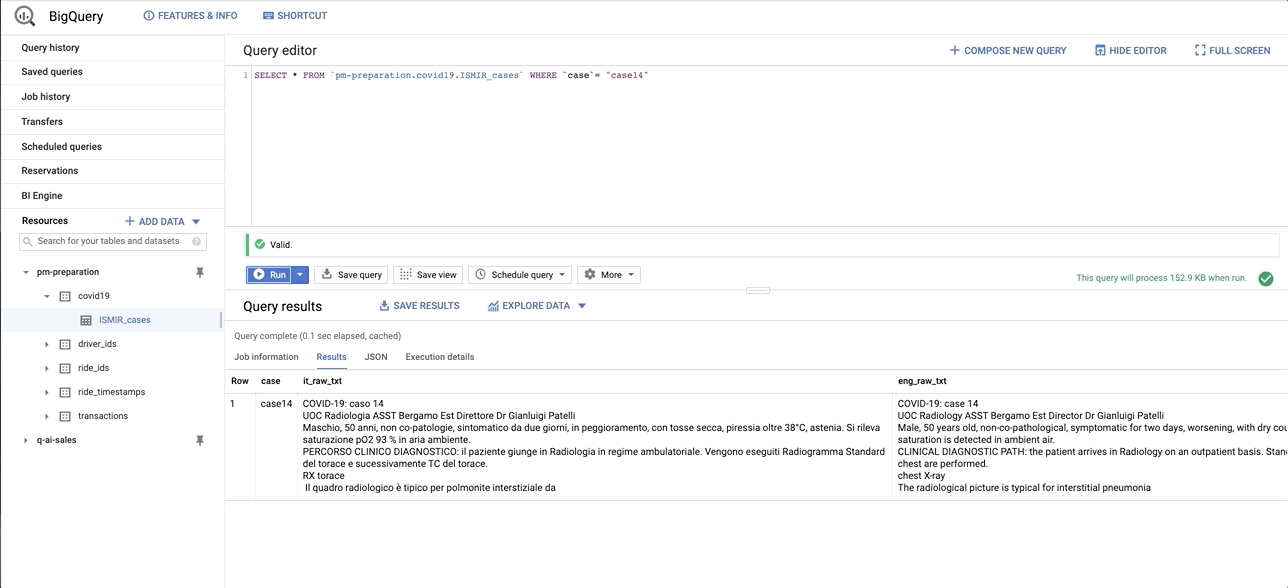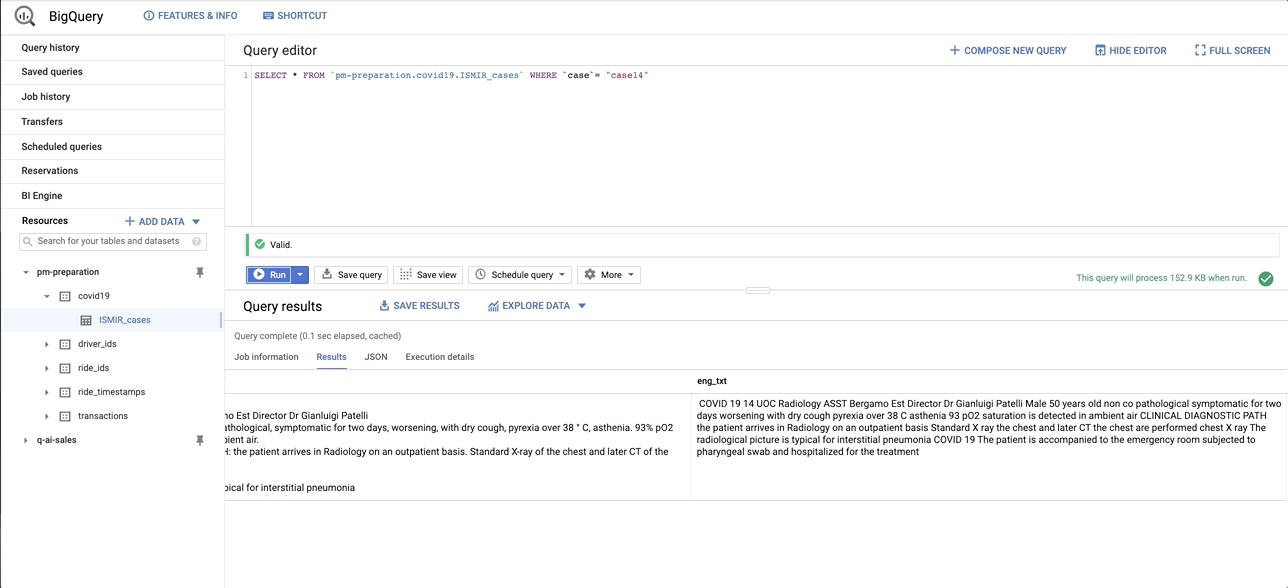
+ 0
- 0
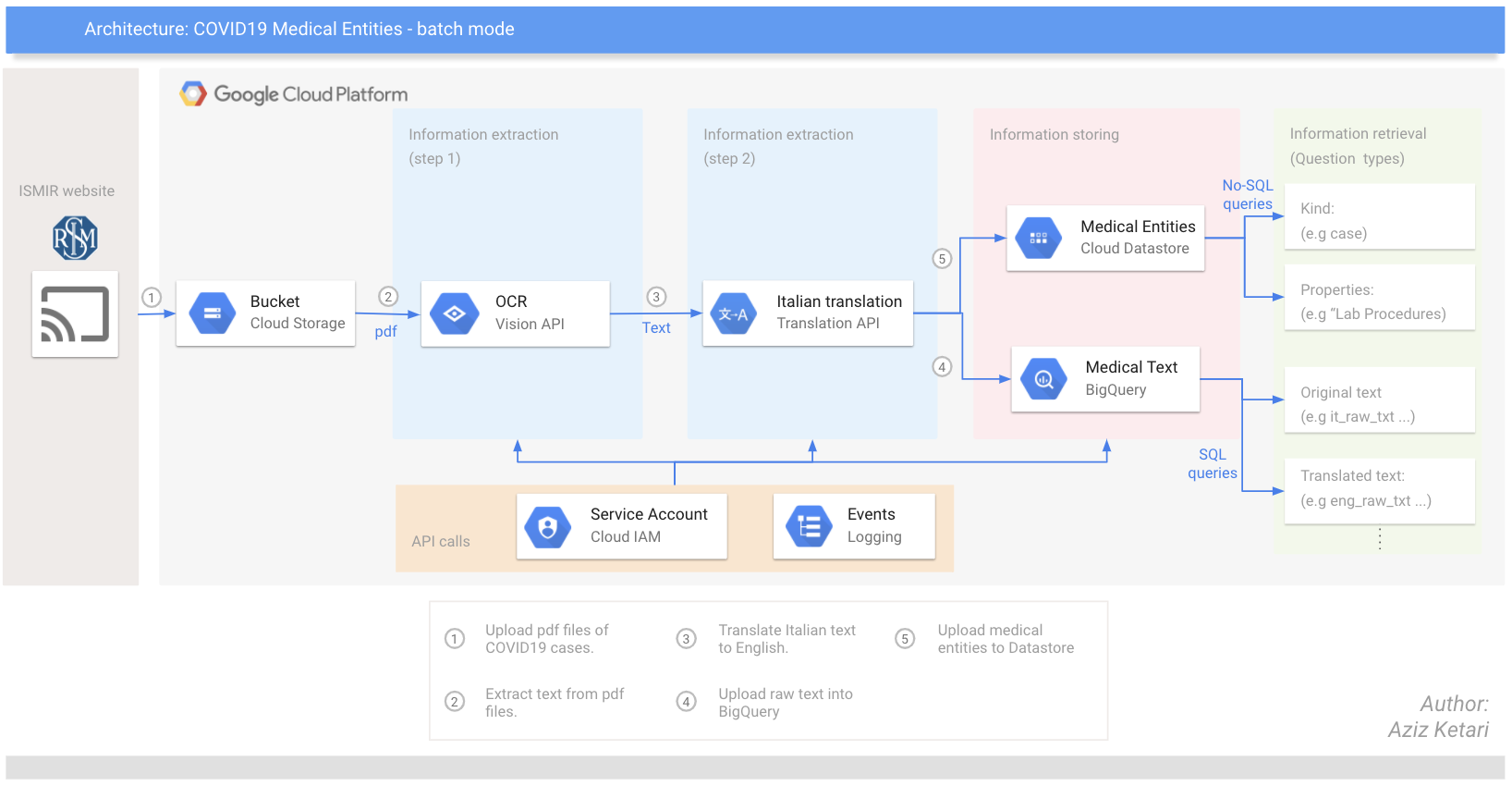
data/images/covid19_repo_architecture_3_24_2020.png → content/images/covid19_repo_architecture_3_24_2020.png
+ 0
- 0
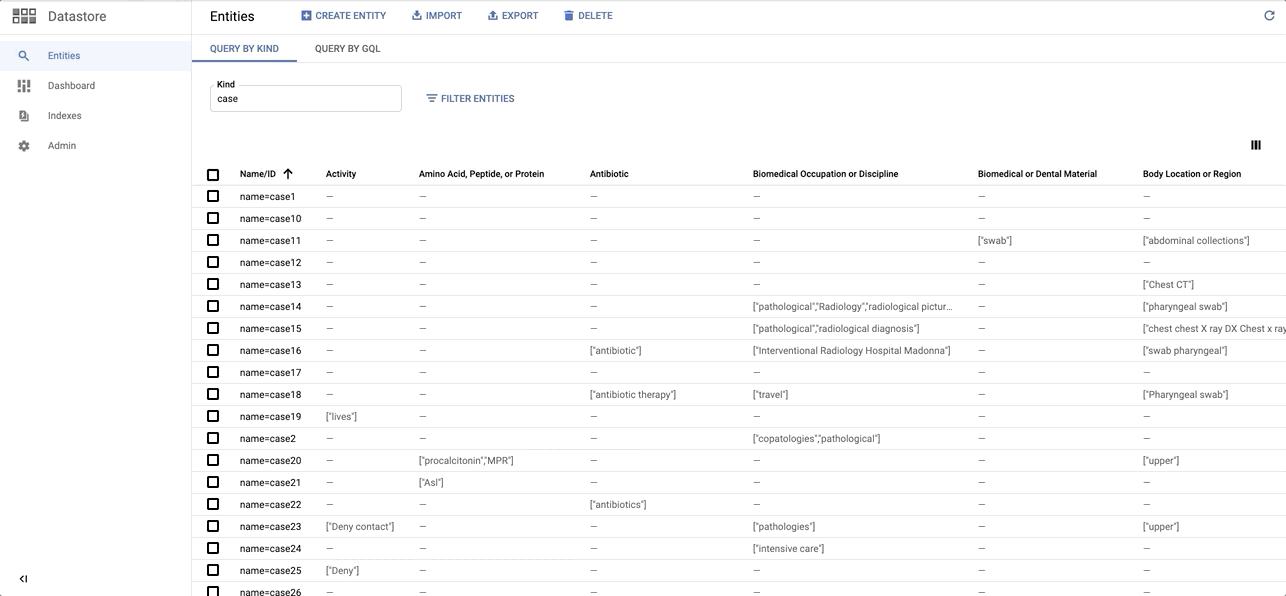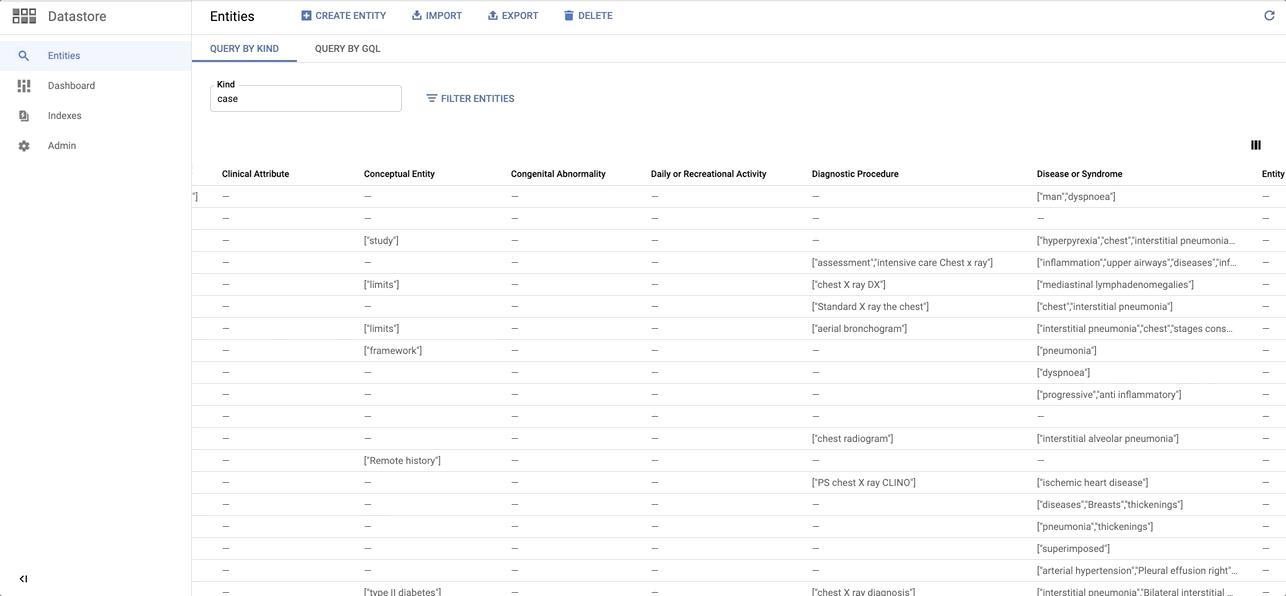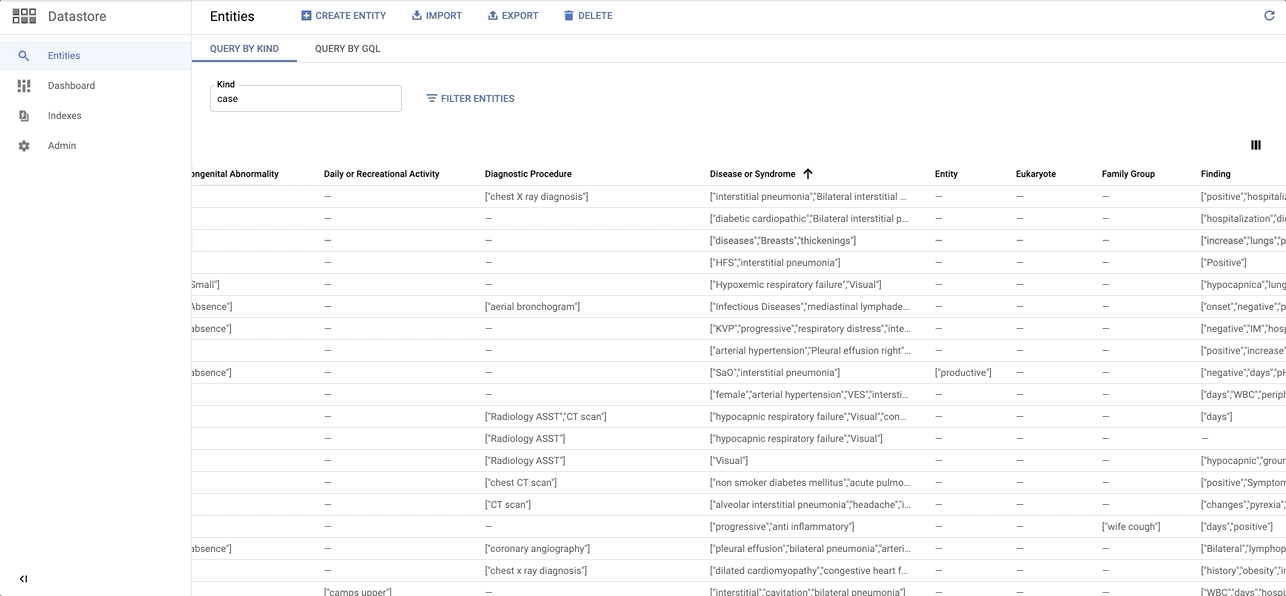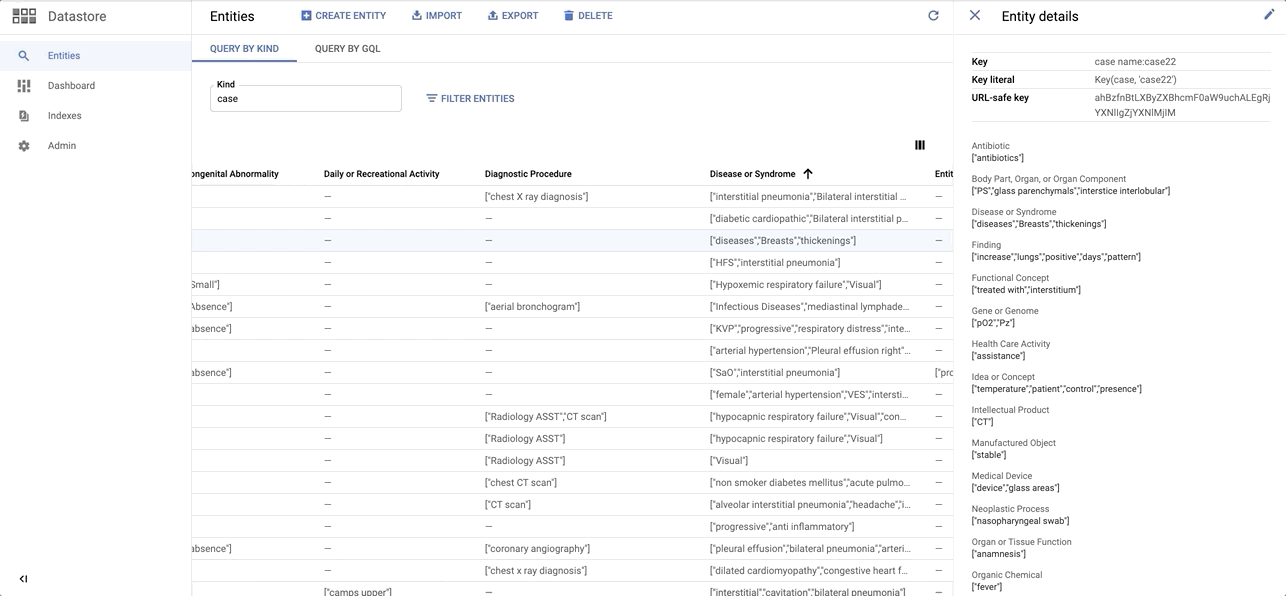
data/images/datastore_snapshot.gif → content/images/datastore_snapshot.gif
+ 0
- 0
scripts/__init__.py
+ 1
- 1
extraction.py → scripts/extraction.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 1
- 1
preprocessing.py → scripts/preprocessing.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 2
- 2
retrieving.py → scripts/retrieving.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 2
- 2
storing.py → scripts/storing.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||